BioCyc contains the following types of regulatory information:
- Regulation of enzymatic activity, usually by small molecules or metal ions. This includes required enzyme cofactors, allosteric activation and inhibition, competitive inhibition, and other forms of enzyme modulation.
- Regulation of gene transcription or translation. This includes regulation of transcription by transcription factors and sigma factors, attenuation, and regulation of translation by small RNAs, riboswitches, and regulatory proteins.
- Regulation by protein modification, such as phosphorylation or binding of a ligand, to determine whether or not a protein is in its active form.
When looking at a gene page, the various regulatory influences on the gene and its product are summarized in the Regulation Summary Diagram at the top of the Summary tab (if this diagram is not present, then there is no regulatory data available for that organism). Mousing over any element of this diagram describes its particular mode of regulatory action. More detailed information is available in the other tabs, as described below.
 |
| Regulation Summary Diagrams for three EcoCyc genes, illustrating a range of regulation types. a) Transcription of trpD is inhibited by TrpR bound to tryptophan; translation of trpD is attenuated by trp-tRNA; the TrpDE enzyme requires Mg2+as a cofactor, and its activity is inhibited by tryptophan. b) Transcription of oppA is inhibited by several different transcription factors, and requires sigma factor σ28; translation of oppA is activated by a spermidine riboswitch and inhibited by small RNA GcvB with accessory protein Hfq. c) Transcription of uvrY is inhibited by LexA; translation of uvrY is activated by regulatory protein DeaD; the protein UvrY is converted to its phosphorylated form by BarA-P. |
Sources of Regulation Data
Regulation of Enzyme Activity
 |
| A portion of the Reactions tab for gene glpK in EcoCyc, showing substrate-level regulation data. |
Regulation of Transcription and Translation
 |
| A portion of the Operons tab for gene fumC in EcoCyc |
Below the Gene Local Context diagram, a separate diagram is shown for each individual transcription unit to clarify which regulatory sites control which promoters. Clicking on one of these diagrams brings up a dedicated transcription unit page providing detailed information for the promoter and each regulator and site, including sequences.
Regulation by Protein Modification
Modified forms of a protein each have their own dedicated page within BioCyc. The attribute table in the Summary tab for a gene page lists and provides links to all alternative forms of a protein. In addition, if there are reactions and pathways that show the interconversion between different forms, those will be included in the Reactions tab (and, briefly, in the page header section). If the location of the protein modification site or binding region is known, it will appear as a feature under the Protein Features tab. For example, compare different sections of the page for ArcB with one of its phosphorylated forms.
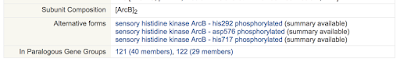 |
| The portion of the Summary tab for gene arcB in EcoCyc that shows the links to alternative forms |
Searching Regulation
The best way to find all entities regulated by some gene or compound is to navigate to the page for that gene or compound, and select the appropriate tab.
The Regulon tab of a gene page shows all transcription units that are regulated by all forms of the gene product.
The Regulation tab of a compound page lists all enzymatic reactions for which the compound is an activator, inhibitor, cofactor, or other regulator. It also shows all transcription units that are regulated by the compound, either directly (e.g. as part of a riboswitch) or as ligand to a transcription factor.
 |
| The Regulation tab for compound spermidine in EcoCyc. |
The command Search → Search Genes, Proteins or RNAs includes an option to search or filter by small molecule regulator, cofactor, substrate or ligand. Although the list of results will be similar to what you could find by visiting the Regulation tab of the compound page, using the dedicated search page allows you to combine this query with other filters. It also outputs the results in the form of a table, which can be converted to a SmartTable.
The command Search → Search DNA or mRNA Sites allows you to search for different types of sites on the chromosome, including transcription units, promoters, terminators, transcription factor binding sites, mRNA binding sites, riboswitches and attenuators. These can be filtered by regulator protein, RNA, or small molecule, as well as by evidence code.
Viewing Regulation in Pathway Pages
By default, pathway diagrams depict regulation at a minimal detail level. A circle icon containing a small plus or minus sign next to an enzyme name means that activation or inhibition data is available for the enzyme or its gene. Moving the mouse over one of these icons generates a tooltip with information about the regulators and type of regulation. In addition, if an enzyme is directly regulated by a small molecule that is a main substrate of the pathway, then a faint gray arrow will point from the substrate to the plus or minus icon.
Clicking on the Show Regulation Details button (this button will be present only if the pathway is displayed at a level of detail that shows enzyme names, and if regulatory data is available for the pathway) will show the pathway diagram at full regulation detail level, including all activators and inhibitors for each enzyme and gene in the pathway (enzyme cofactors and sigma factors are not included). Moving the mouse over any regulator generates a tooltip with more details.
 |
| The pathway diagram for L-homoserine biosynthesis in EcoCyc, with regulation details shown. |
A pathway page also includes transcription unit diagrams for all of the pathway's genes, showing transcription factor and other regulatory binding sites. The Genetic Regulation Schematic shows both direct and indirect transcriptional and translational regulators.
The Regulatory Overview
The Regulatory Overview diagram shows all genes that either are transcription factors or sigma factors, or are regulated by transcription factors or sigma factors. Users can interactively select connections to be displayed. This diagram is only available if the currently selected organism is one for which transcriptional regulation data exists. For more information about how to interact with this diagram, see the Website User Guide.
 |
| The EcoCyc Regulatory Overview Diagram, before showing any connections. |

No comments:
Post a Comment