The Metabolic Route Search Problem
Consider the problem of performing an in-depth exploration of the metabolic network of an organism that you study, to compare alternative paths within that network whereby the organism can transform a starting metabolite into an ending metabolite. What are the lengths and properties of these alternative pathways?Consider now a broader problem, namely the metabolic-engineering problem of finding the most efficient modification to the biochemical network of an organism to allow the organism to synthesize a new metabolite from a feedstock compound. One aspect of "most efficient" is minimize the number of reactions added from an external database of known reactions.
RouteSearch [1] is a Pathway Tools component that solves both of the preceding problems by computing optimal metabolic routes, that is, an optimal series of biochemical reactions that connects start and goal compounds, given various cost parameters to control the optimality of the routes found. RouteSearch can display several of the best routes it finds using an interactive graphical web page. When RouteSearch is used for metabolic engineering, it uses the MetaCyc database as its external reaction database.
In computing optimality, RouteSearch takes into account the conservation of nonhydrogen atoms from the start compound to the goal compound. Perhaps surprisingly, it is possible to devise reaction paths that conserve no atoms from start to goal compound! The more atoms that are conserved, the more efficient the transformation from start to goal. To compute the number of conserved atoms, RouteSearch uses precomputed atom mappings of reactions that are available in MetaCyc [2]. An atom mapping of a reaction gives a one to one correspondence of each nonhydrogen atom from reactants to products.
RouteSearch is available only in Web mode in Pathway Tools (since version 17.0, March 2013). It is also available at BioCyc.org but without the possibility to add reactions from MetaCyc (that mode is available only for locally installed versions of Pathway Tools). More details on how to use MetaCyc with RouteSearch are given in the following section.
Using the RouteSearch Software
In the following we will describe RouteSearch and how to use it at BioCyc.org. The first step is to point your browser to BioCyc.org. RouteSearch is currently supported for the Firefox and Chrome browsers whereas the Safari and Internet Explorer browsers are not supported by RouteSearch due to their inability to completely support SVG graphics.Once at biocyc.org, select your preferred organism by clicking the "change organism database" link in the right-side box of the Web page (unless the current organism next to "Searching" is already your preferred organism). For RouteSearch, any database can be selected except MetaCyc. Then, select the command Metabolism -> Metabolic Route Search from the top menubar. A Web page similar to the following will be shown (here for E. coli):
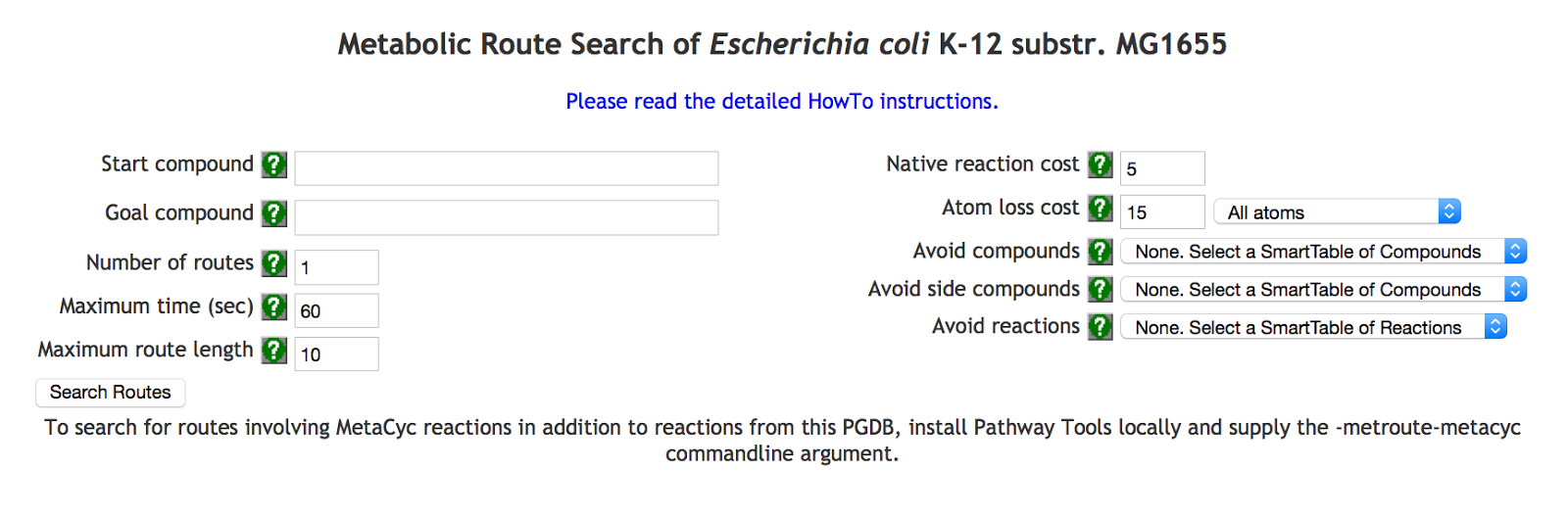
From now on, the selected organism is the prefered source of reactions to use to find metabolic routes. We call these reactions, the native reactions. Before searching for routes, some parameters need to be given and others can be changed. Default values are already provided for most parameters and they can be left as is for now. Online documentation is provided by clicking the question marks next to each parameter name in the Web interface of RouteSearch. For each parameter, an entry box, or selector, is provided. The parameters are:
- Start and Goal compounds of the routes to find. Once you start typing a compound name (or frame identifier) in the boxes next to these parameters, an auto-completion mechanism will help you select the desired compound.
- Number of routes to find. We recommend to first use 1 to make the tool run faster. We limit the number of routes to find so that it is reasonable to search and display them.
- Maximum time (in seconds) to use to find the routes. If the maxium time is used by RouteSearch, the best solutions found before running out of time are displayed but these routes could be suboptimal, they are not necessarily optimal routes.
- Maximum route length. If this parameter is greater than about 14, RouteSearch could take a long time to find any route.
- Allow use of MetaCyc reactions? When MetaCyc is allowed, reactions from MetaCyc can be used by RouteSearch to construct metabolic routes, and compounds not existing in the selected organism but existing in MetaCyc can be specified as start and goal compounds. This parameter is not available at BioCyc.org; to allow MetaCyc, see the following section.
- Cost of using one reaction from MetaCyc in a route. This cost is enabled only when MetaCyc is allowed. Each reaction from MetaCyc added to the route will increase the cost of the route by this specified amount.
- Cost of using one reaction from the selected organism in a route. Each reaction from the selected organism added to the route will increase the cost of the route by this specified amount.
- Atom lost cost and list of atom species (as a selector). These parameters allow you to select the atom species to take into account when lost in a route and the cost of losing one such atom from the start to the goal compound in a route. When you select "All atoms", all atom species except hydrogen atoms are account for. If you select "Selected atom species", the list of atom species displayed will be "C O P N S". The box can be edited by clicking in it and typing. You can remove some atom species, for example, by deleting all letters except "C" that specifies the carbon atoms. At least one space must separate atom species (i.e., letters). The other atoms are oxygen (O), phosphate (P), nitrate (N), and sulfur (S). Currently, no other atom species can be specified.
- Finally, you can select compounds and reactions not to include in any routes to find, or even side compounds that cannot be used in any reactions of the routes to find.
Once all fields are filled, you click the "Search Routes" button. The request is sent to the Web server to compute the optimal routes. Once found, the optimal routes are returned and displayed on the Web page under the parameters. The list of displayed routes are sorted in increasing cost.
For example, let us select the organism E. coli, define the start compound as succinate, the goal compound as (S)-malate, and all other fields left as is (i.e., default values). This example is very simple since the TCA cycle of E. coli has a short route from succinate to (s)-malate, but the short route obtained allows its complete display on this Web page. RouteSearch found the following route:
In this particular route, eight atoms were kept from source to target compound, only two reactions are used, and its cost is 10 (i.e., two reactions multiplied by cost five, no atoms lost). In general, each route is numbered, and for each route, the number of atoms kept from the start to the goal compound, its number of reactions and cost are displayed on the left. The route itself is shown by displaying each compound structure, from the start to the goal compound, colored according to the moieties conserved along the route. Between each compound, an arrow is shown representing a reaction, and under each arrow the name of the enzyme catalyzing that reaction. If an arrow is red, it represents a reaction from MetaCyc, otherwise it is gray and it represents a reaction from the selected database. Clicking the arrow opens up a new browser tab to display the reaction page.
Note that since the routes can be long, and that computer screen monitors are limited in size, it is likely that the Web page needs to be horizontally scrolled to see the right part of the routes.
Mousing over any atom of the compounds will highlight the atom along the route from compound to compound. This highlighting allows us to follow the route of an atom and discover if it reaches the target compound, and if it does not reach it, which reaction loses it along the route.
For a more complex example, involving new reactions added from MetaCyc, please see the next section.
Enabling MetaCyc Searching by RouteSearch within your Pathway Tools Web server
You need to run Pathway Tools locally on your computer to enable inclusion of MetaCyc reactions in routes computed by RouteSearch. Starting Pathway Tools in Web mode with MetaCyc enabled for RouteSearch is done with the command line option "-metroute-metacyc", such as:
ptools -www -metroute-metacyc
When accessing the Metabolic Route Search Web page of your Web server, via a browser as described in the previous section, you will get a Web page similar to the following. In particular, notice that the parameter "Allow use of MetaCyc reactions?" is present which is due to the option "-metroute-metacyc" specified on the command line above.
Please consult the Pathway Tools User Guide for more information on how to start Pathway Tools in Web server mode. The User Guide is only available as part of the Pathway Tools software distribution.
The following screen snapshot shows the interface and result once a search for routes from succinate to ethylene was launched with MetaCyc searches enabled. We set the cost of using reactions from MetaCyc to 10, that is, two times greater than the cost of a native reaction from E. coli. The route keeps two carbon atoms from succinate to ethylene. This is the maximum possible since ethylene has only two carbon atoms. Notice that the last reaction in the route found is red: this means that this reaction is from MetaCyc. The other two reactions are from E. coli.
For even more complex examples please see paper [1].
[1] Mario Latendresse, Markus Krummenacker, and Peter D. Karp.
Optimal Metabolic Route Search Based on Atom Mappings,
Bioinformatics, doi: 10.1093/bioinformatics/btu150, March 2014.
[2] Mario Latendresse, Jeremiah P. Malerich, Mike Travers, and Peter D. Karp.
Accurate Atom-Mapping Computation for Biochemical Reactions,
Journal of Chemical Information and Modeling (JCIM), September 2012.


No comments:
Post a Comment