BioCyc contains gene essentiality datasets for 30 organisms (see listing at end). These datasets have been both collected from individual publications, and harvested in bulk from the OGEE database (see
http://ogee.medgenius.info/browse/). In some cases, multiple essentiality datasets are available for a given organism (e.g., for
E. coli K-12).
BioCyc essentiality datasets are keyed to the conditions of growth (e.g., MOPS medium with 0.4% glucose), since different sets of genes will be essential under different conditions of growth. If essentiality data is available for a given gene, it will be shown on the gene page, under the essentiality tab.
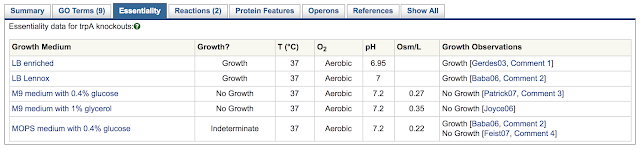 |
| The Essentiality tab for gene trpA in EcoCyc |
In addition, BioCyc provides access to the full lists of essential and non-essential genes for a given dataset. To obtain such lists:
- Select your organism of interest using "change organism database" in the upper right corner.
- Click Analysis menu → Summary Statistics
- In the bottom table, the table will contain an entry "Gene Essentiality Datasets" if any essentiality datasets are present. Click on those words.
- In the resulting page, click on the entry in the "Growth Medium" column for the dataset of interest.
- The resulting pages will show lists of genes that are essential and/or non-essential for growth on this medium. To manipulate those gene lists, click "Turn into a SmartTable". SmartTable results can be downloaded as a file and explored interactively.
Current BioCyc organisms with gene essentiality data are as follows. To generate the list:
- Click "change organism database" in upper right corner of page
- Click "By Organism Properties" tab
- In "Select Property" selector, select "# Genes with Essentiality Data"
- Change "has values" selector to "has any value"
- Click "Find Organisms"
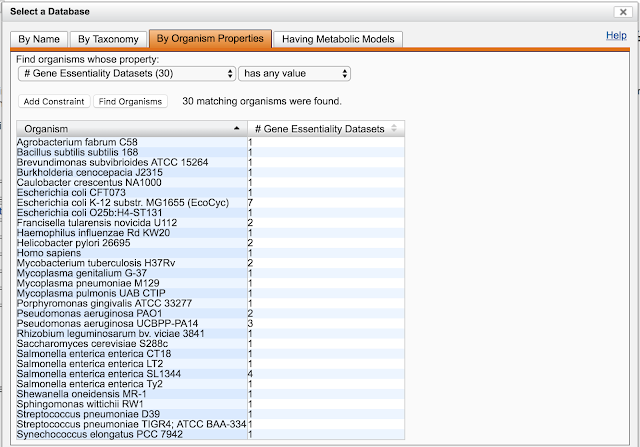 |
| The organism selector showing all databases with gene essentiality datasets |
- Agrobacterium fabrum C58
- Bacillus subtilis subtilis 168
- Brevundimonas subvibrioides ATCC 15264
- Burkholderia cenocepacia J2315
- Caulobacter crescentus NA1000
- Escherichia coli CFT073
- Escherichia coli K-12 substr. MG1655 (EcoCyc)
- Escherichia coli O25b:H4-ST131
- Francisella tularensis novicida U112
- Haemophilus influenzae Rd KW20
- Helicobacter pylori 26695
- Homo sapiens
- Mycobacterium tuberculosis H37Rv
- Mycoplasma genitalium G-37
- Mycoplasma pneumoniae M129
- Mycoplasma pulmonis UAB CTIP
- Porphyromonas gingivalis ATCC 33277
- Pseudomonas aeruginosa PAO1
- Pseudomonas aeruginosa UCBPP-PA14
- Rhizobium leguminosarum bv. viciae 3841
- Saccharomyces cerevisiae S288c
- Salmonella enterica enterica CT18
- Salmonella enterica enterica LT2
- Salmonella enterica enterica SL1344
- Salmonella enterica enterica Ty2
- Shewanella oneidensis MR-1
- Sphingomonas wittichii RW1
- Streptococcus pneumoniae D39
- Streptococcus pneumoniae TIGR4; ATCC BAA-334
- Synechococcus elongatus PCC 7942



No comments:
Post a Comment