BioCyc contains the following types of regulatory information:
- Regulation of enzymatic activity, usually by small molecules or metal ions. This includes required enzyme cofactors, allosteric activation and inhibition, competitive inhibition, and other forms of enzyme modulation.
- Regulation of gene transcription or translation. This includes regulation of transcription by transcription factors and sigma factors, attenuation, and regulation of translation by small RNAs, riboswitches, and regulatory proteins.
- Regulation by protein modification, such as phosphorylation or binding of a ligand, to determine whether or not a protein is in its active form.
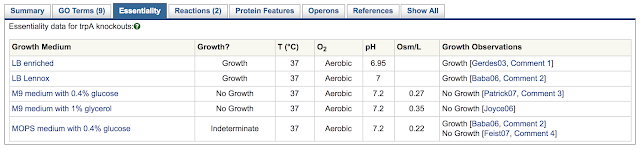
When looking at a gene page, the various regulatory influences on the gene and its product are summarized in the Regulation Summary Diagram at the top of the Summary tab (if this diagram is not present, then there is no regulatory data available for that organism). Mousing over any element of this diagram describes its particular mode of regulatory action. More detailed information is available in the other tabs, as described below.
 |
| Regulation Summary Diagrams for three EcoCyc genes, illustrating a range of regulation types. a) Transcription of trpD is inhibited by TrpR bound to tryptophan; translation of trpD is attenuated by trp-tRNA; the TrpDE enzyme requires Mg2+as a cofactor, and its activity is inhibited by tryptophan. b) Transcription of oppA is inhibited by several different transcription factors, and requires sigma factor σ28; translation of oppA is activated by a spermidine riboswitch and inhibited by small RNA GcvB with accessory protein Hfq. c) Transcription of uvrY is inhibited by LexA; translation of uvrY is activated by regulatory protein DeaD; the protein UvrY is converted to its phosphorylated form by BarA-P. |